Simplify Machine Learning Pipeline Analysis with Object Storage
Machine Learning is bringing exciting opportunities for applications in healthcare and will be a key enabler of digital medicine and new discoveries. Many organizations are looking at how they can adopt Machine Learning for research and analysis. In this blog post I will walk you through a standard Machine Learning pipeline, some of the challenges in adoption and how you can build a better Machine Learning pipeline analysis using on-premises object storage.
[Tweet “Collaboration + privacy + scale –> A better #machinelearning pipeline for research. #BioIT18”]
Data In, Intelligence Out
Machine Learning pipelines consist of several steps to train a model. The term “pipeline” is misleading as it implies a one-way flow of data. However, Machine Learning pipelines are cyclical. Every step is repeated to continuously improve the accuracy of the model, until an adequate algorithm is achieved.
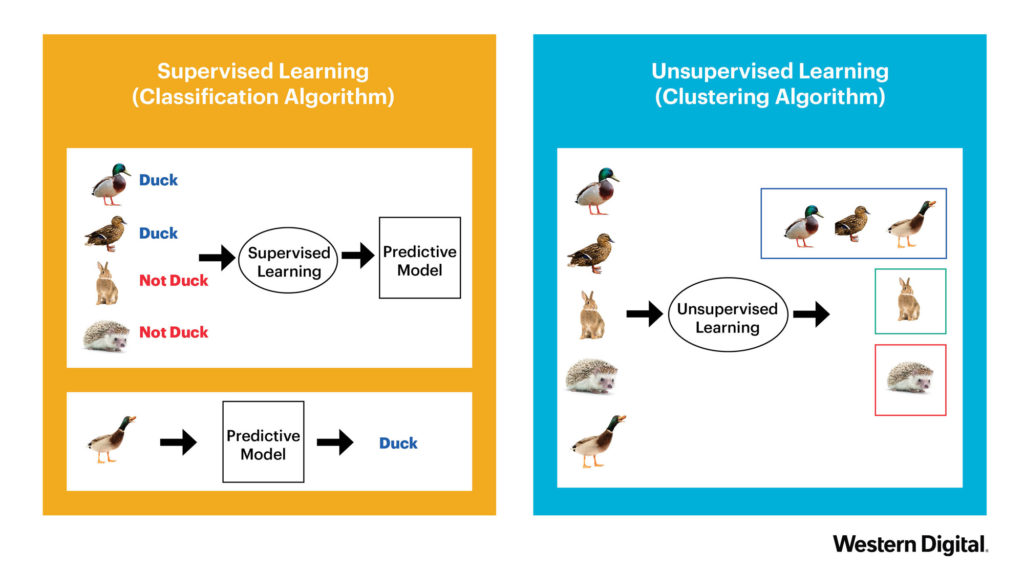
There are generally two types of learning approaches. The first is Supervised Learning. This is what you probably associate with Machine Learning. A model is built by providing a dataset with concrete example. (e.g. this is a duck, this is not a duck). The second approach, Unsupervised Learning, is used to discover structures within given data. The initial data is not necessarily labeled and the learning uses clustering algorithms in order to group unlabeled data together. See the example below.
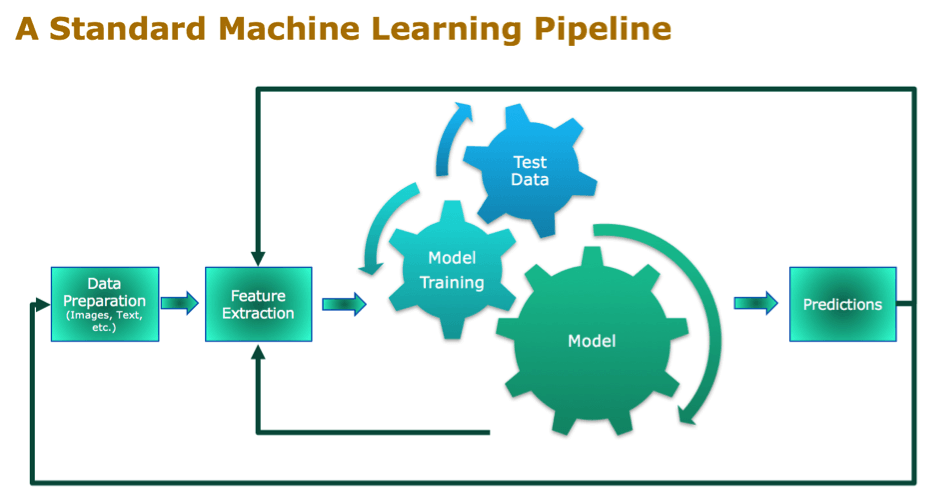
Standard Machine Learning Pipeline
One of the first steps in the Machine Learning pipeline is feature selection or extraction. You want to reduce the dimensionality of the data as a preprocessing step and remove any irrelevant and redundant data. This not only saves processing power, time and costs, but it will also significantly increase the accuracy and comprehensibility of your model.
Feature Extraction
One way to do this is using feature selection (or attribute selection). This is the selection of attributes, or a subset of features, that will be most relevant in creating a predictive model. The second is feature extraction, which is the extraction of existing features and their transformation into a new format that describes the variance within the data and reduces the amount of information needed to represent it. As this involves transformation of the features, including losing certain information in the process, it is often not reversible. You cannot recreate the original image from the extracted features.
Feature extraction is particularly interesting in the field of health and medical research and analysis for reasons of data privacy. Using this method, private patient information and image source cannot be shared or recreated once they enter the pipeline.
Single Source of Truth (SSOT)
Another key element of Machine Learning pipeline in the context of research is having a single repository with the single source of truth. You want the source data to be in a single repository so that you can point to as a reference. Using this architecture you can run Machine Learning on the data from various points or locations, and not have to carry or port it to whatever location the analysis is being done at. This helps to avoid duplicate and varying versions, replicated values being forgotten, and makes sure multiple teams, and even multiple institutions, are always working with the single truth of data.
Petabyte-Scale Architecture Using Object Storage Private Cloud
One of the big challenges that anyone working in Life Sciences confronts every day is scale. Bioimaging technologies are advancing rapidly. Techniques such as 4D require over a terabyte of data per scan. With such a demanding data volume, many researchers suddenly find that not only do they have budget and storage capacity problems, but they also need to become IT experts in order to solve it!
Thankfully, now there’s object storage. Object storage is scalable by design and supports highly concurrent access. It is extremely resilient and it’s built to be cost-effective. (Not familiar with object storage? Read about it here) Furthermore, it enables reproducible Machine Learning pipelines with object versioning.
Machine Learning Pipeline Example:
Running NVIDIA® Deep Learning GPU Training System (DIGITS) Using the ActiveScaleTM System
For this example pipeline I used Western Digital’s ActiveScale object storage system, a turnkey, petascale solution with Amazon S3™ compatibility, and NVIDIA DIGITS.
NVIDIA DIGITS provides a graphic user interface via a web browser for users. This provides a great advantage over many code-based applications, by opening access to Machine Learning to more scientists and end users. The current supported frameworks are NVCaffe™, Torch, and TensorFlow™.
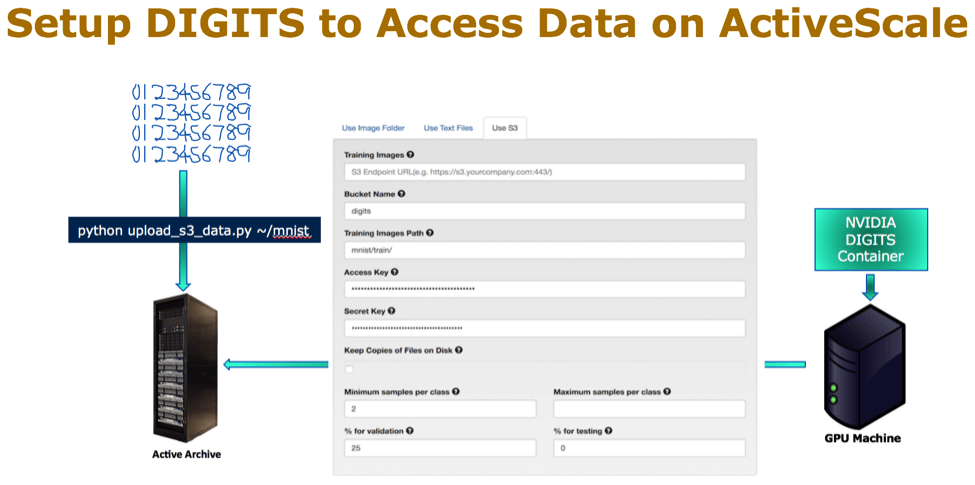
In this example, I used the DIGITS container deployment with single GPU machine and the MNIST data as a test example. The MNIST database of handwritten digits has a training set of 60,000 examples. DIGITS container provides Python® scripts to upload data to an S3 storage target. Using the provided scripts I uploaded the MNIST data to the ActiveScale object bucket.
ActiveScale supports the S3 protocol, and can be used as a target in the same way a public cloud would be. Then, I configured from the DIGITS S3 setting page to point to the ActiveScale bucket. The following picture illustrates these steps:
After setting up DIGITS for MNIST, I chose the classification algorithm to create a classification dataset. Now, I am ready to train the model with the LeNet model. Once I have the predictive model, I can classify test data. In this example, I used the digit “9”, which the model took 6 seconds to classify the test image.
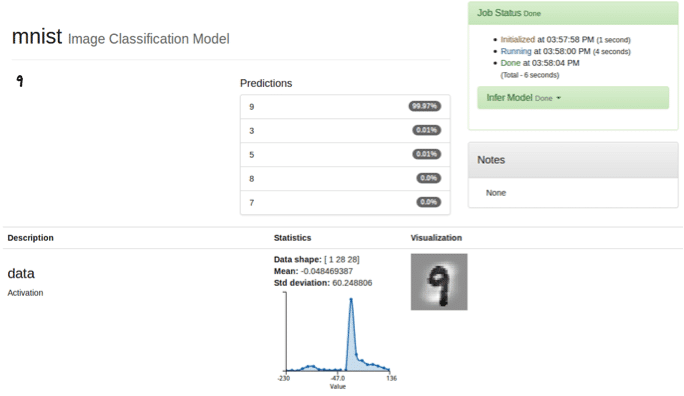
As you can see from the above steps, it is really easy to run training on DIGITS with ActiveScale. One can deploy DIGITS on different machines and reproduce the same training by pointing DIGITS to the ActiveScale bucket with the initial data we uploaded for this training. ActiveScale is the single truth of data repository for our machine learning pipeline.
Bringing it All Together – Feature Extraction, Data Privacy, SSOT, Scale
With this single repository on ActiveScale, you can take the DIGITS container and point the library to run the same experiment from anywhere. You can run analysis without needing to upload the data again or replicate it. Using S3 you can even point public cloud compute resources to this one dataset and remote train around the world.
Furthermore, with feature extraction you keep on premise control of the original data. That data, let’s say raw images that contain patient information, is kept on premises in the ActiveScale system. Only the feature-extracted data runs in the cloud.
Data Preservation – a Future of Learning
Machine Learning and AI will drive the future of research and medicine. Accessible, scalable, and durable storage solutions are the foundation for breakthroughs in life sciences. Make sure you understand what steps your organization needs to take to leverage these new advancements.
Join us at Bio-IT world in booth #501 to learn more, or visit our research and life sciences solutions page.
Forward-Looking Statements
Certain blog and other posts on this website may contain forward-looking statements, including statements relating to expectations for our product portfolio, the market for our products, and the capacities, capabilities and applications of our products. These forward-looking statements are subject to risks and uncertainties that could cause actual results to differ materially from those expressed in the forward-looking statements, including development challenges or delays, changes in markets, demand, global economic conditions and other risks and uncertainties listed in Western Digital Corporation’s most recent quarterly and annual reports filed with the Securities and Exchange Commission, to which your attention is directed. Readers are cautioned not to place undue reliance on these forward-looking statements and we undertake no obligation to update these forward-looking statements to reflect subsequent events or circumstances.